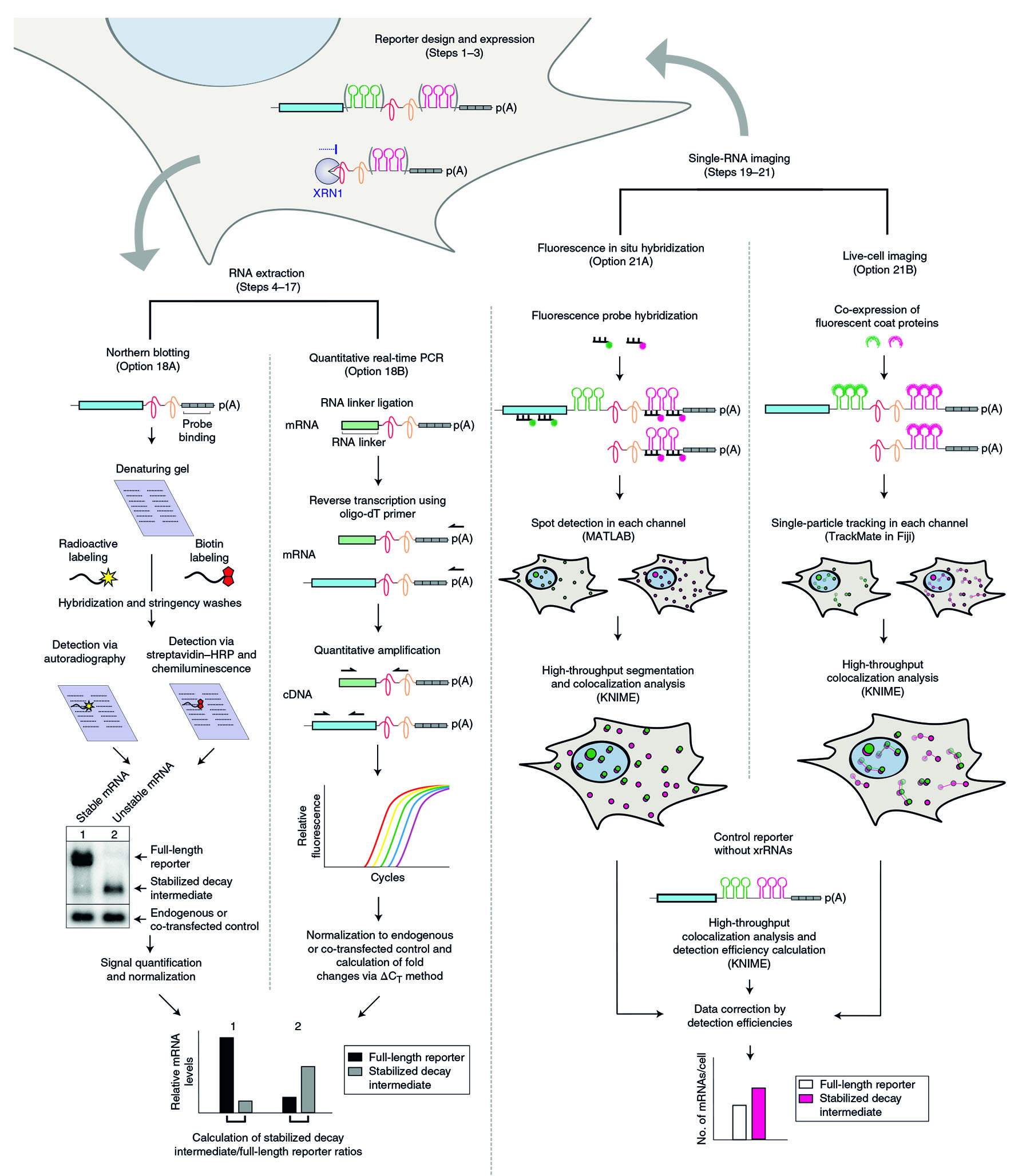
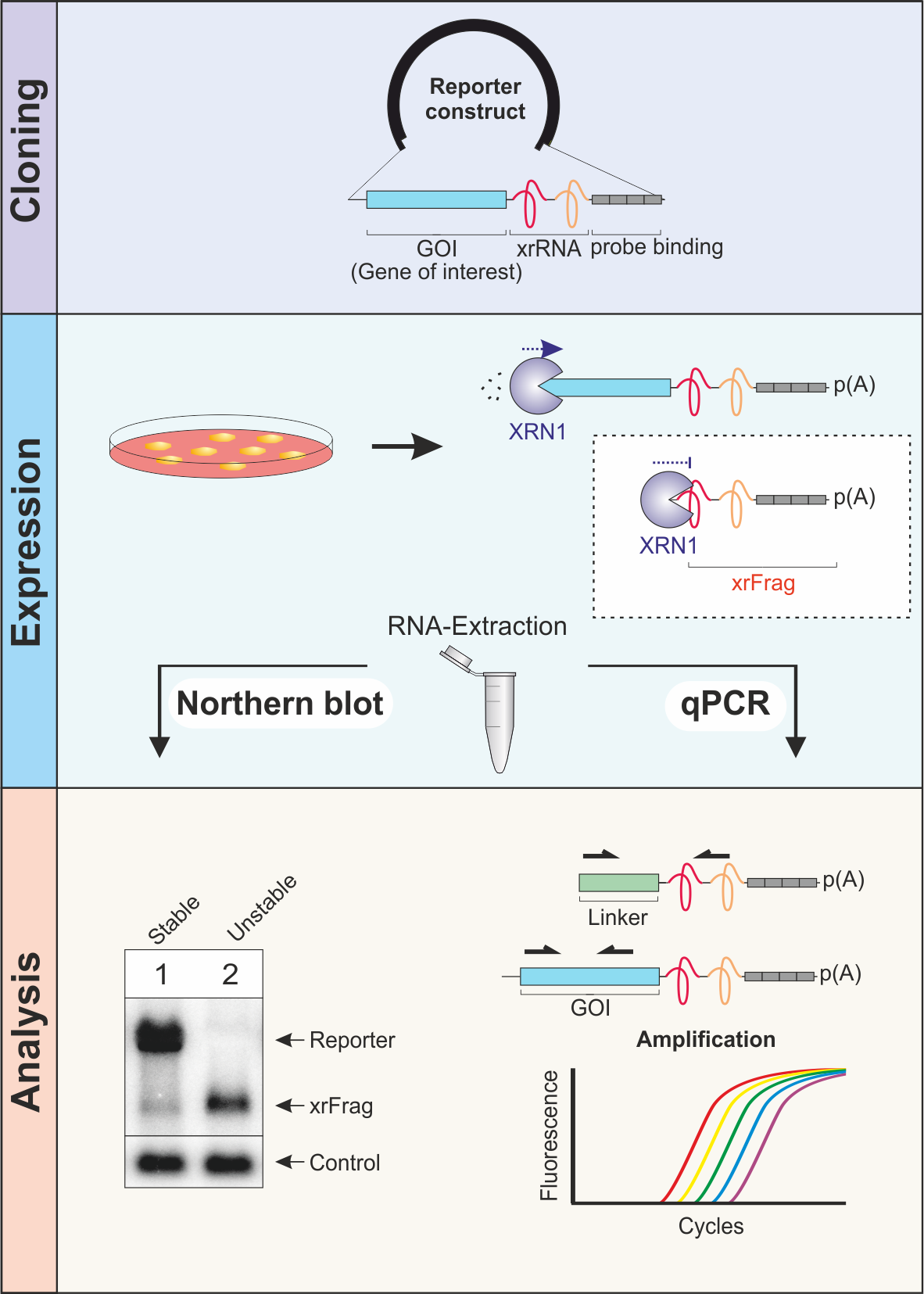
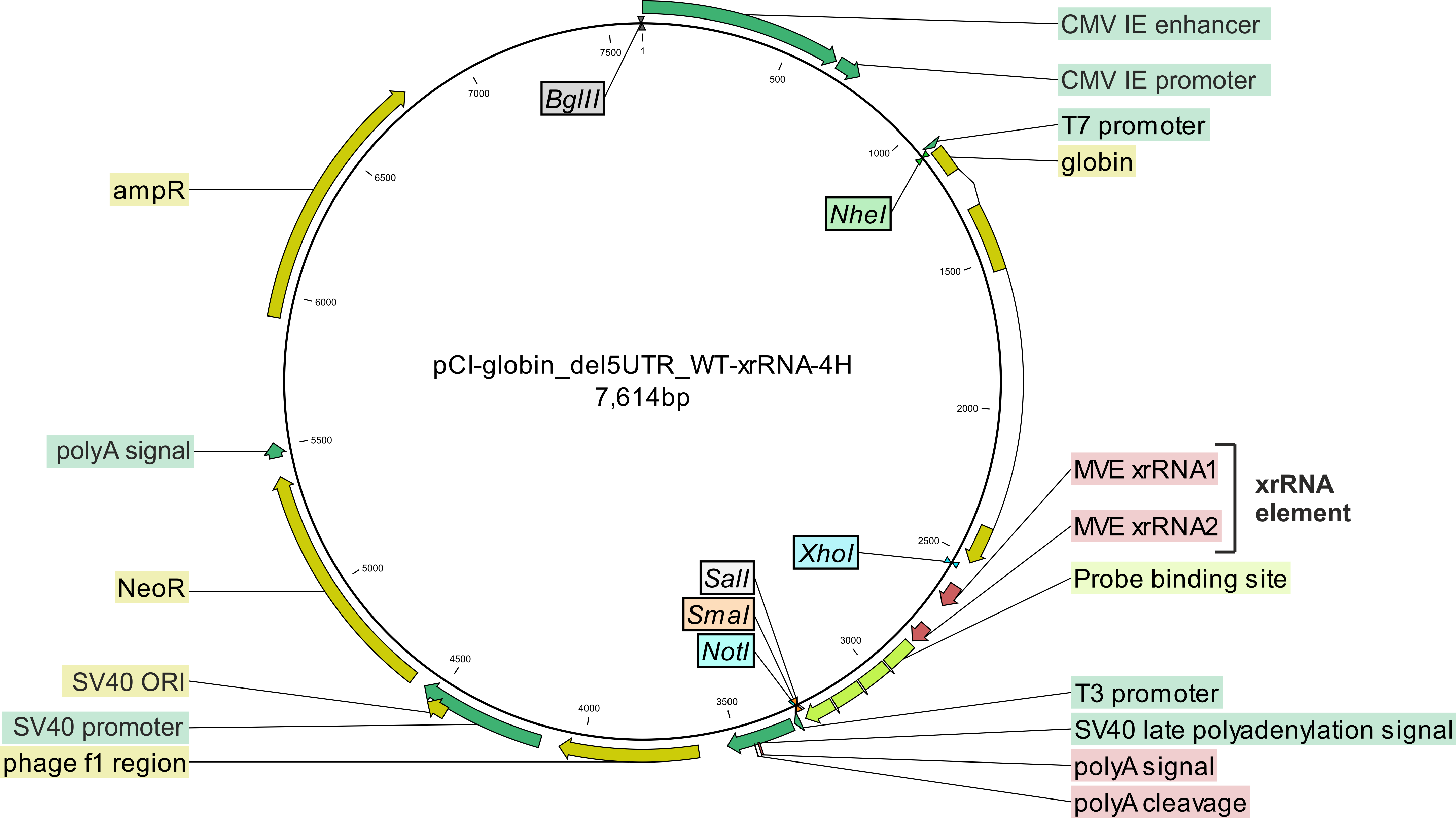
The xrFrag method
All cellular transcripts are generally degraded by the contribution of at least one of the typical decay pathways depicted below, namely exonucleolytic or endonucleolytic degradation.
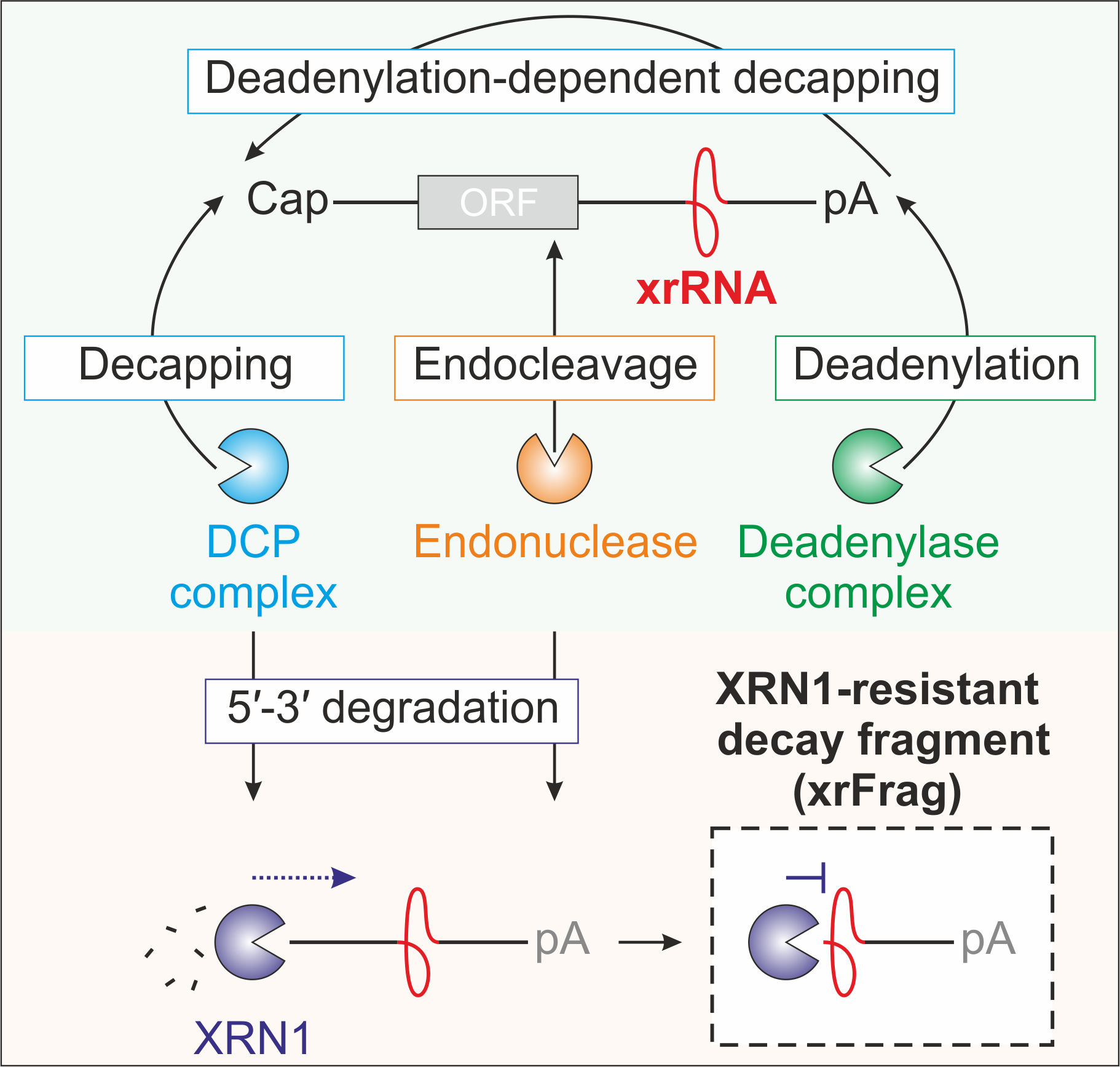
Image from Boehm et al. 2016 Nat Comm
In order to identify the pathway that is responsible for the turnover of a certain transcript of interest, we developed the xrFrag method (xrFrag = XRN1-resistant decay fragment).
Using the virus-derived xrRNA element, decay intermediates that arise due to the turnover of RNAs can be visualized.
We have successfully used the xrFrag method to analyze multiple mRNA decay pathways such as nonsense-mediated mRNA decay (NMD), miRNA-induced decay and degradation caused by AU-rich elements or cytokine 3' UTRs
If you are interested, here you can find the original publication: PubMed