Projects > Splicing in Physiology, Aging and Disease
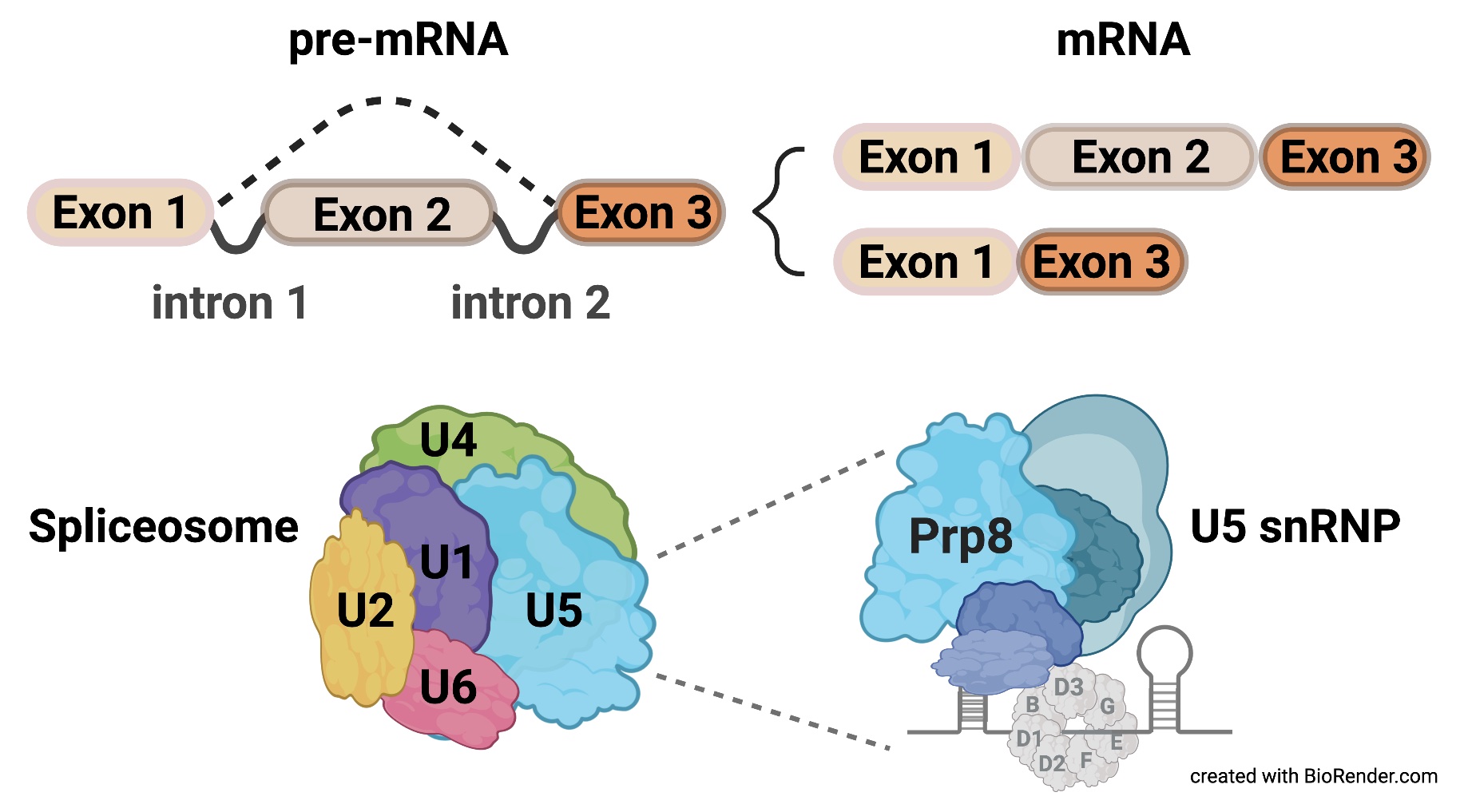
Most protein-coding genes in higher eukaryotes are discontinuous, containing introns that interrupt the amino acid-coding sequences called exons. To generate translation-competent mature mRNA molecules, introns must be excised, and exons joined in a process known as pre-RNA splicing. Pre-mRNA splicing represents a crucial step in the regulation of gene expression pivotal to tissue and organ development, homeostasis, and organismal healthspan. The “cut and join” process catalyzed by a spliceosome must be efficient and precise but also flexible to generate specific transcripts on demand. In contrast, mutations in, and deregulation of, spliceosome components underlie numerous human diseases and organismal aging. It remains puzzling how malfunctions in core spliceosome components manifest in tissue-specific pathogenesis rather than a systemic disease.
We study how spliceosome assembles and functions in physiology, stress, and disease. We are interested in unraveling the mechanisms underlying spliceosome plasticity, tissue-specific pre-mRNA splicing, and differential sensitivity of cells to splicing aberrations to help the development of novel splicing modulators/inhibitors with therapeutic potential.
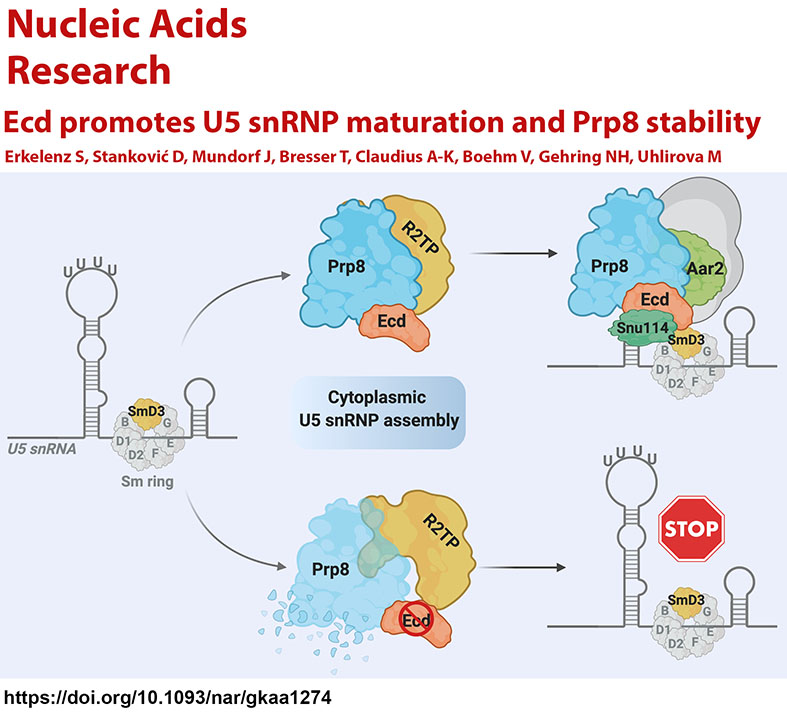
Exploiting the genetic amenability of the Drosophila system, we established an in vivo animal model to study the mechanisms underlying both the etiology and pathology of Retinitis pigmentosa type 13 caused by mutations in Prp8, the largest and the most conserved protein of the U5 snRNP of the spliceosome (Stankovic et al., 2020). We identified an evolutionarily conserved protein Ecdysoneless (Ecd) as a novel interactor of Prp8 (Claudius et al., 2014) and demonstrated that Ecd functions as a critical mediator of U5 snRNPs assembly. We showed that Ecd guards Prp8 stability and facilitates its delivery to the forming U5 snRNP particles in the cytoplasm via interactions with the Sm ring protein SmD3 and U5 snRNA. Without Ecd, Prp8 is destabilized, and U5 snRNP biogenesis is stalled, resulting in genome-wide changes in gene expression and splicing (Erkelenz et al., 2021).