Problems Inherent in the Species Concepts and Possible Work-Arounds
- Shortcomings of the Different Species Concepts
- Incongruences Between Species Concepts and their Consequences
- How to Cope with the Species Problem?
- DNA Barcoding
Shortcomings of the Different Species Concepts
Most protistan morphospecies have been described from field samples, i.e. a researcher observed one, several or many – sometimes fixed – cells displaying the same morphology and described them as a new species. Intra-clonal or intra-specific variation of morphological characters cannot be safely addressed in field material. Also it is usually impossible to identify character states that have been passed on by an ancestor to several, but not all descendent evolutionary lineages. Therefore ancestral character states may be misleading. If such a character state has been used for delineating taxa, the result will be an inconsistent systematics with an unnatural classification into groups. Also heteromorphic life histories of protists usually cannot be identified. Different life stages of the same species therefore have and had been erroneously described as separate species. Most heteromorphic life histories, but also intra-specific variation have been discovered only in laboratory cultures, whereas plesiomorphic (ancestral) character states can be identified only by molecular phylogenetic analyses (Pringsheim 1955; Hill and Wetherbee 1986; Fresnel 1994; Hoef-Emden and Melkonian 2003).
The biological species concept cannot be applied, if organisms reproduce only asexually, if sexual reproduction is not known or if mating experiments are not possible. In such cases an alternative species concept has to be chosen. Usually the morphospecies concept has been the fallback option.
A DNA-based taxonomy such as the phylogenetic species concept highly depends in the variability of the chosen DNA marker and in the chosen threshold of genetic divergence between two species (Evans et al. 2007). The more conserved a DNA sequence is, the less species will be recognised, the higher variable a DNA sequence, the higher the species counts.
Although CBCs in the internal transcribed spacer 2 seem to be reliable indicators for non-interbreedability, a CBC clade may still encompass several biological species and thus, may require later revision. Besides, application of the CBC clade concept is more difficult than simply defining species by phylogeny, by a molecular signature or by morphology. First of all, the ribosomal operon repeats of a eukaryote may contain several versions of ITS2, whereas the rRNA genes are usually identical. Even in a clonal culture indels comprising several nucleotides may be present in the less conserved parts of the different ITS2 versions. Thus, ITS2 sequences often cannot be sequenced directly from the PCR product, but require cloning. Secondary structure prediction of ITS2 sequences usually does not work right out-of-the-box. The ITS2 sequences have to be submitted in pieces that correspond to the helical domains to the RNA folding server. To be able to identify the domains, an alignment of several ITS2 sequences of closely related taxa are required. Servers providing software pipelines for secondary structure prediction may help to facilitate the procedure (Schultz and Wolf 2009).
Incongruences Between Species Concepts and their Consequences
 If enough reliable and distinctive morphological
characters are available, morphological, phylogenetic and biological species may be congruent.
Severe problems in systematics occur, if morphological characters either do not reflect genetic
diversity or are grossly misleading (see above).
Especially microscopic eukaryotes have become
notorious for systematic problems. Many protistan morphospecies have been described in the past, but a
lack of reliable and/or distinctive morphological characters resulted
in so-called cryptic species complexes (e.g. Mann et al. 2004; Hoef-Emden 2007; Lilly et al. 2007; Ujiie and
Lipps 2009). In other cases, cells from the same clonal culture, but also cells from other cultures that are
genetically identical in the chosen molecular marker, display different cell types that correspond to different
morphospecies (Hoef-Emden 2007).
If enough reliable and distinctive morphological
characters are available, morphological, phylogenetic and biological species may be congruent.
Severe problems in systematics occur, if morphological characters either do not reflect genetic
diversity or are grossly misleading (see above).
Especially microscopic eukaryotes have become
notorious for systematic problems. Many protistan morphospecies have been described in the past, but a
lack of reliable and/or distinctive morphological characters resulted
in so-called cryptic species complexes (e.g. Mann et al. 2004; Hoef-Emden 2007; Lilly et al. 2007; Ujiie and
Lipps 2009). In other cases, cells from the same clonal culture, but also cells from other cultures that are
genetically identical in the chosen molecular marker, display different cell types that correspond to different
morphospecies (Hoef-Emden 2007).
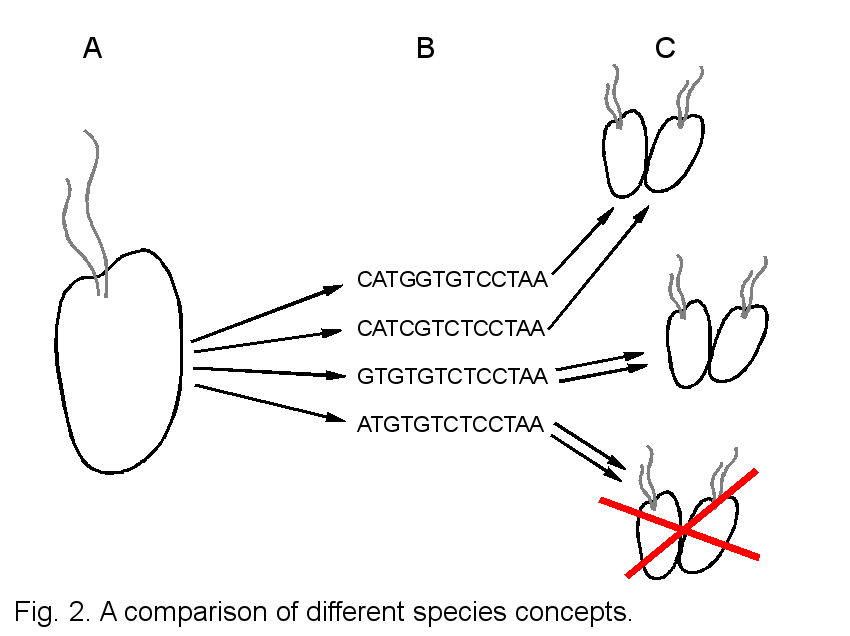
Possible relationships between a cryptic morphospecies complex and other species concepts are depicted in Fig. 2 (2A, morphospecies; 2B, phylogenetic species; 2C, biological species). Morphologically similar to identical cells (2A) may be genetically very different. Under a phylogenetic species concept or a DNA-based taxonomy, morphology can be neglected and each genotype is assigned a separate species name (2B). Depending in the resolution of chosen molecular markers and thresholds of divergence, however, two genetically different individuals may be sexual compatible and produce viable and fertile offspring = represent one biological species (2C, top). In this case the chosen molecular marker has been too variable or the threshold of genetic divergence to distinguish between species has been set too low. If phylogenetically defined species interbreed only within cells of the same genotype, the choice of molecular marker and threshold have been optimal and correlate exactly with biological species (2C, middle). In a third scenario, two cells of the same genotype may be incompatible, indicating that the chosen molecular marker has been too conserved to resolve between biological species (2C, bottom).
Since species counts depend in the applied species concept and/or chosen molecular markers, biodiversity surveys may yield extremely different results for the same habitat. Surveys using the morphospecies concept often result in lower species counts, than surveys based on environmental DNA. Biological species cannot be assigned at all in biodiversity surveys, because this requires clonal cultures and time-consuming mating experiments. Besides. sexual reproduction or inductors of sexual reproduction are not even known in most protists.
How to Cope with the Species Problem?
Sometimes a detailed reexamination of morphological characters and molecular phylogenetic analysis unveil that suitable diagnostic characters are present, but have been overlooked before. Some diatoms proved to be such pseudo-cryptic species (Mann et al. 2004). Since mating experiments have been possible in these diatoms, the congruence of morphological, DNA-based and biological species could be tested. In other protistan groups, morphological characters fail to resolve the genetic diversity behind. In addition, the congruence of DNA-based species with biological species often cannot be confimed. Under such circumstances, using the CBC clade concept to define species probably will result in species approaching putative biological species as close as currently possible. The CBC clade concept can be combined with phylogenetic analyses using more conserved phylogenetic markers. The ITS2 sequences are necessary to identify putative biological species, whereas conserved phylogenetic markers are necessary to infer inter-specific relationships at a higher resolution. Although CBC clades may encompass several biological species, they represent clearly defined, reproducible and non-interbreedable taxonomic units. If in future a possibility to determine biological species will be found in the respective group, this will probably not turn the CBC clade taxonomy upside down in a way that biological species boundaries cross CBC clades. It may be just necessary to split CBC clades into as many species as required and complementing the CBC clades by additional means.
Revision of a protistan group with ambiguous morphospecies has to follow the rules of the respective nomenclature code to be valid. The oldest fitting species description always has priority over later ones. A taxonomist, who wants to revise a group, has to decide which name should be assigned to a species. In most protists the holotype is of no help, since it is, different from land plants or animals, usually a more or less accurate drawing or – in diatoms – an empty silica shell devoid of organic material. Under these circumstances, it is impossible to isolate DNA and to add a molecular signature to an ambiguous species description. Thus a taxonomist has the choice of either saving old species names by arbitrarily linking them to molecular signatures/CBC clades of a morphologically cryptic species complex or to neglect the names as ambiguous and describe all CBC clades as new species. In the latter case, the old species names will become useless historical waste.
The current versions of zoological and botanical nomenclature codes are almost completely focused on morphology. Both nomenclature codes insist on voucher material. In protistan species voucher materials are usally microscopic slides or preparations for electron microscopy, despite the fact that in many protists these holotypes may be as futile as drawings for identification. The botanical code of nomenclature has been adopted to some extend, by allowing cultures of fungi or algae preserved "in a metabolically inactive state" (i.e. cryopreserved or lyophylised) to be used as holotypes. Unfortunately, many protists don't survive cryopreservation or lyophilisation procedures. To avoid problems with identification of a described species in future, it is therefore a good option to always submit a DNA sample of the newly described species together with the holotype to a publicly accessible collection and to mention it in the text accompanying the species diagnosis.
DNA Barcoding
DNA barcoding aims at identifying species with short and highly variable DNA sequences. Since taxonomists have become rare, such short DNA barcodes could facilitate an identification of species without expertise (Barcode of Life Initiative). A preferred barcode marker for animals is the 5' terminus of the subunit I of the mitochondrial cytochrome oxidase c gene (cox1 or COI, around 600 to 800 nucleotides) (Hebert et al. 2003). This DNA sequence for sure, cannot be used in amitochondriate taxa. Also it proved to be too conserved to barcode embryophyte plants. Therefore a set of other DNA barcode markers have been proposed for the Embryophyta (Hollingsworth et al. 2009). As central hubs for the growing masses of data, publicly accessible databases have been designed. They are supposed to link literature, morphological and geographical data and one or several DNA barcodes to each species (e.g. Barcode of Life Database [BOLD]). Such databases thus represent attempts to integrate morphological, biological and DNA-based species concepts.
To serve well as a barcode, a DNA sequence has to be highly variable and short, which contradicts the requirements for a good phylogenetic marker, which is supposed to resolve also deeper divergences. Thus, if a group of organisms is largely undersampled, a blastn search with a DNA barcode such as COI against a nucleotide database may yield completely misleading results even on class level. If DNA barcoding is applied to a group with an irreproducible species concept (cryptic species complexes, para- and/or polyphyletic taxa), the results of a clustering analysis will reflect the systematic confusion. Under such circumstances it will not be possible to assign names of morphospecies to clusters of DNA barcode sequences. Meyer and Paulay have demonstrated that DNA barcoding works only in well-characterised and reasonably sampled groups (Meyer and Paulay 2005). Thus to establish an accurately working DNA barcode system, an often cumbersome morphological reexamination in combination with phylogenetic analyses and a subsequent revision of protistan groups is inevitable.
Thanks to the progress in DNA sequencing techniques, masses of DNA barcodes can be generated within a short time from environmental DNA. However, many of these DNA sequences neither can be assigned to a morphology nor to a species name, resulting in huge lists of "faceless phantoms" in the sequence databases. This is not exactly a satisfying situation for a biologist. Considering the overwhelming diversity of microscopic organisms, revising all the protistan groups is a huge task requiring not only many taxonomists, but also a lot of time. Strategies to solve the taxonomic problems and to speed up this process are in work (Hoef-Emden et al. 2007).